This post describes the steps for building and deploying a machine learning model for heart disease prediction. From dataset exploration and feature engineering to model training, evaluation, and Docker deployment, this blog post covers the essential steps in the machine learning lifecycle, providing practical insights and code examples along the way.
Step 1: Load & Explore Dataset
The Python code below imports the necessary libraries and proceeds to load the heart disease dataset, which will serve as the foundation for the content in this blog post.
# Import Libraries
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
import seaborn as sns
############################################
# STEP 1: IMPORTING DATA
############################################
# Specify the path to your CSV file
file_path = '/Users/heart_disease_df_1.csv'
# Load the CSV file into a DataFrame
heart_disease_df = pd.read_csv(file_path)Step 2: Exploratory Data Analysis (EDA)
Exploratory Data Analysis is a preliminary phase in data analysis where the focus is on understanding the main characteristics of a dataset, uncovering patterns, and identifying trends using statistical and visual methods.
The provided Python code initially displays all column names along with the count of non-null values and data types. Subsequently, it utilizes the Seaborn library to create visualizations illustrating the distribution of age, the correlation between age and serum cholesterol level, the maximum heart rate categorized by gender, and the distribution of chest pain types.
############################################
# STEP 2: EXPLORATORY DATA ANALYSIS (EDA)
############################################
# Print information about the DataFrame
print(heart_disease_df.info())
----------------- OUTPUT -----------------------
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 age 1076 non-null int64 # Age of the individual
1 sex 1076 non-null int64 # Gender of the individual (0 = female, 1 = male)
2 cp 1023 non-null float64 # Chest pain type
3 trestbps 1076 non-null int64 # Resting blood presure
4 chol 1021 non-null float64 # Serum cholesterol level
5 fbs 1076 non-null int64 # Fasting blood sugar
6 restecg 1028 non-null float64 # Resting electrocardiographic results
7 thalach 1076 non-null int64 # Maximum heart rate achieved
8 exang 1076 non-null int64 # Exercise-induced angina
9 oldpeak 0 non-null float64 # ST depression induced by exercise relative to rest
10 slope 1076 non-null int64 # Slope of the peak exercise ST segment
11 ca 1076 non-null int64 # Number of major vessels colored by fluoroscopy
12 thal 1076 non-null int64 # Thalassemia
13 target 1076 non-null int64 # Target variable indicating the presence or absence of heart disease (0 = no heart disease, 1 = heart disease)
------------------------------------------------
# Set the style for Seaborn
sns.set(style="whitegrid")
# Create visualizations with Seaborn
plt.figure(figsize=(12, 6))
# Distribution of Age
plt.subplot(2, 2, 1)
sns.histplot(df['age'], kde=True)
plt.title('Distribution of Age')
# Age vs Cholesterol
plt.subplot(2, 2, 2)
sns.scatterplot(x='age', y='chol', hue='target', data=df)
plt.title('Age vs Cholesterol')
# Gender vs Max Heart Rate
plt.subplot(2, 2, 3)
sns.boxplot(x='sex', y='thalach', data=df)
plt.title('Gender vs Max Heart Rate')
# Chest Pain Type Distribution
plt.subplot(2, 2, 4)
sns.countplot(x='cp', hue='target', data=df)
plt.title('Chest Pain Type Distribution')
plt.tight_layout()
plt.show()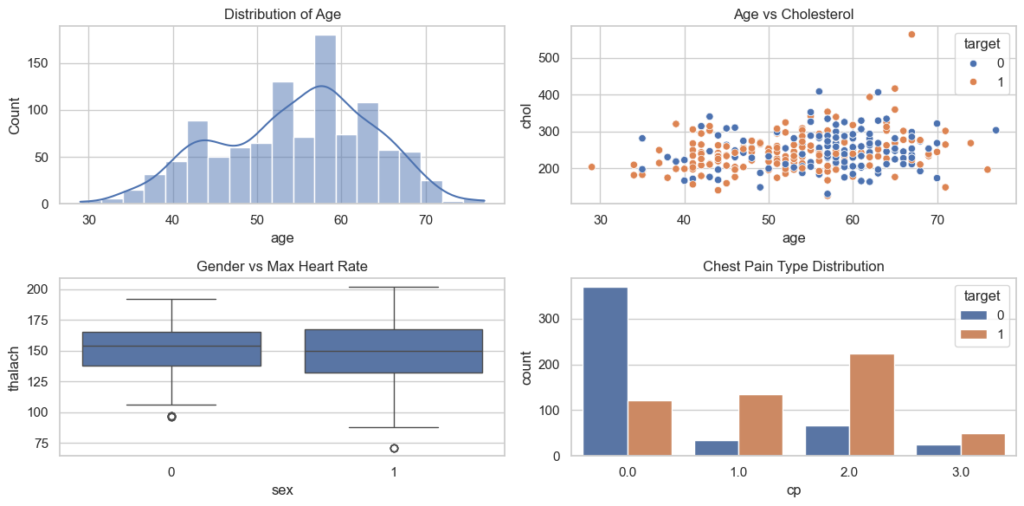
The visualizations above illustrate the age distribution, the correlation between age and serum cholesterol level, the maximum heart rate categorized by gender, and the distribution of chest pain types. As observed in the bottom right corner, a notable insight is that if the Chest Pain Type is greater than 0.0, there is a higher likelihood of heart disease compared to non-heart disease.
Step 3: Data Preparation
Data preparation is a crucial step in the data analysis process, involving the cleaning, transforming, and organizing of raw data into a format suitable for analysis.
The Python script presented below eliminates the ‘oldpeak’ column from the data frame due to its absence of values. Following this, an assessment is conducted to identify columns containing null values, revealing that cp, chol, and restecg exhibit such gaps. Given the categorical nature of cp and restecg, the script proceeds to remove rows with null values in these columns. Concurrently, it fills null values in the chol column with the mean value.
############################################
# STEP 3: DATA PREPARATION
############################################
# Remove the 'oldpeak' column from the DataFrame
heart_disease_df = heart_disease_df.drop(['oldpeak'], axis=1)
# Check and display the number of null values in each column
heart_disease_df.isnull().sum()
----------------- OUTPUT -----------------------
age 0
sex 0
cp 53
trestbps 0
chol 55
fbs 0
restecg 48
thalach 0
exang 0
slope 0
ca 0
thal 0
target 0
dtype: int64
------------------------------------------------
# Remove rows with null values in the 'cp' and 'restecg' columns
heart_disease_df = heart_disease_df.dropna(subset=['cp'])
heart_disease_df = heart_disease_df.dropna(subset=['restecg'])
# Calculate the mean value of the 'chol' column
mean_value = heart_disease_df['chol'].mean()
# Impute missing values in the 'chol' column with the mean
heart_disease_df['chol'].fillna(mean_value, inplace=True)
# Check and display the number of null values in each column after data preparation
heart_disease_df.isnull().sum()
----------------- OUTPUT -----------------------
age 0
sex 0
cp 0
trestbps 0
chol 0
fbs 0
restecg 0
thalach 0
exang 0
slope 0
ca 0
thal 0
target 0
dtype: int64
------------------------------------------------Step 4: Feature Engineering
Feature engineering is the process of creating new, meaningful features or transforming existing ones in a dataset to enhance the performance and interpretability of machine learning models.
Normalization:
- One common feature engineering techniques is normalization. Normalization scales numeric features to a scale of 0 to 1 ensuring that no particular feature can dominate the model due to its scale. This is beneficial when features have different ranges and you use algorithms sensitive to the input’s scale like K-Nearest Neighbors (KKN) or Neural Networks.
Standardization:
- Another common feature engineering techniques is standardization. Standardization scales feature to have a mean of zero and a variance of one. Standardization benefits algorithms that assume features are centered around zero and have variance in the same order, like in Support Vector Machines (SVMs) and Linear Regression.
In the Python code below, we will be using standardization for the numerical fields to scale features to have a mean of zero and a variance of one.
############################################
# STEP 4: FEATURE ENGINEERING
############################################
from sklearn.preprocessing import StandardScaler
# Extract numerical columns (excluding 'target') for standardization
numerical_columns = heart_disease_df.select_dtypes(include=np.number).columns.drop('target')
# Create a StandardScaler instance
scaler = StandardScaler()
# Standardize the numerical columns
heart_disease_df[numerical_columns] = scaler.fit_transform(heart_disease_df[numerical_columns])
# Display the standardized DataFrame
print(heart_disease_df.head())
----------------- OUTPUT -----------------------
age sex cp trestbps chol fbs restecg \
0 -0.284493 0.667283 -0.928778 -0.379466 -0.674237 -0.429235 0.904416
1 -0.173932 0.667283 -0.928778 0.481382 -0.857711 2.329728 -0.991753
2 1.705598 0.667283 -0.928778 0.768331 -1.448905 -0.429235 0.904416
3 0.710553 0.667283 -0.928778 0.940501 -0.857711 -0.429235 0.904416
4 0.821113 -1.498615 -0.928778 0.366602 0.997416 2.329728 0.904416
thalach exang slope ca thal target
0 0.820547 -0.684962 0.969993 1.181855 1.094562 0
1 0.244504 1.459935 -2.253420 -0.736205 1.094562 0
2 -1.084826 1.459935 -2.253420 -0.736205 1.094562 0
3 0.510370 -0.684962 0.969993 0.222825 1.094562 0
4 -1.926735 -0.684962 -0.641713 2.140885 -0.525721 0
------------------------------------------------Step 5: Feature Selection
Feature selection is a process in machine learning where a subset of relevant and significant features (variables) is chosen from the original set of features. The goal is to improve model performance, reduce overfitting, and enhance interpretability by focusing on the most important attributes.
RandomForestClassifier can be used for feature selection inherently through the way it constructs decision trees. Random Forest is an ensemble learning method that builds multiple decision trees and combines their predictions. During the construction of each tree, a subset of features is randomly selected for consideration at each split.
############################################
# STEP 5: FEATURE SELECTION
############################################
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestClassifier
from sklearn.feature_selection import SelectFromModel
# Define features (X) and target variable (y)
X = heart_disease_df.drop('target', axis=1)
y = heart_disease_df['target']
# Split the data into training and testing sets
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
# Define the random forest model and fit to the training data
rf = RandomForestClassifier(n_jobs=-1, class_weight='balanced', max_depth=5)
rf.fit(X_train, y_train)
# Define the feature selection object
model = SelectFromModel(rf, prefit=True)
# Transform the training features
X_train_transformed = model.transform(X_train)
original_features = X.columns
print(f"Original features: {original_features}")
----------------- OUTPUT -----------------------
Original features: Index(['age', 'sex', 'cp', 'trestbps', 'chol', 'fbs', 'restecg', 'thalach','exang', 'slope', 'ca', 'thal'], dtype='object')
------------------------------------------------
# Select the features deemed important by SelectFromModel
features_bool = model.get_support()
selected_features = original_features[features_bool]
print(f"\nSelected features: {selected_features}")
----------------- OUTPUT -----------------------
Selected features: Index(['age', 'cp', 'thalach', 'exang', 'ca', 'thal'], dtype='object')
------------------------------------------------
feature_importance = pd.DataFrame({
"feature": selected_features,
"importance": rf.feature_importances_[features_bool]
})
plt.figure(figsize=(10, 6))
plt.barh(feature_importance["feature"], feature_importance["importance"])
plt.show()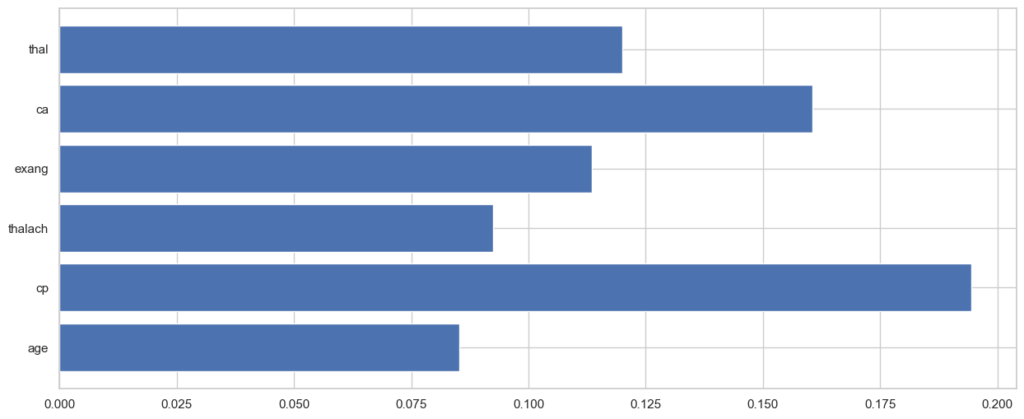
As seen in the chart above, the feature that has the highest importance to heart disease is cp (chest pain type), followed by ca (number of major vessels colored by fluoroscopy) and thal (thalassemia).
Step 6: Model Training & Prediction
In this section, we will use logistic regression to train and predict if a patient has a heart disease. Logistic regression is a supervised machine learning algorithm used for binary classification problems, predicting whether an instance belongs to one of two classes.
Then Python code below imports the necessary libraries from the scikit-learn module. LogisticRegression is employed for creating the logistic regression model, and metrics such as accuracy, classification report, and confusion matrix are assessed using functions from sklearn.metrics.
An instance of the logistic regression model (log_reg_model) is initialized. Logistic regression is a commonly used algorithm for binary classification tasks. The logistic regression model is trained using the fit method, where it learns the patterns and relationships within the transformed training features (X_train_transformed) and their corresponding labels (y_train).
Once the model is trained, predictions are generated on the original test features (X_test). Note that there seems to be a typo in the code (model.transform), and it should likely be log_reg_model.transform. The accuracy score, confusion matrix, and a detailed classification report are computed to evaluate the performance of the trained logistic regression model on the test set (y_test and X_test_transformed).
############################################
# STEP 6: MODEL TRAINING & PREDICTION
############################################
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import accuracy_score, classification_report, confusion_matrix
# Create a logistic regression model
log_reg_model = LogisticRegression()
# Train the model on the transformed training features
log_reg_model.fit(X_train_transformed, y_train)
# Make predictions on the original test features
X_test_transformed = model.transform(X_test)
y_pred = log_reg_model.predict(X_test_transformed)
# Evaluate the model performance
accuracy = accuracy_score(y_test, y_pred)
conf_matrix = confusion_matrix(y_test, y_pred)
classification_rep = classification_report(y_test, y_pred)
# Display model performance metrics
print(f"Accuracy: {accuracy:.2f}")
print("\nConfusion Matrix:\n", conf_matrix)
print("\nClassification Report:\n", classification_rep)
----------------- OUTPUT -----------------------
Accuracy: 0.78
Confusion Matrix:
[[70 23]
[21 82]]
Classification Report:
precision recall f1-score support
0 0.77 0.75 0.76 93
1 0.78 0.80 0.79 103
accuracy 0.78 196
macro avg 0.78 0.77 0.77 196
weighted avg 0.78 0.78 0.78 196
------------------------------------------------
The model’s performance metrics are reported as follows:
- Accuracy: 0.78:
- The accuracy metric indicates that the model correctly predicted the class labels for approximately 78% of the instances in the test set.
- Confusion Matrix:
- The confusion matrix provides a detailed breakdown of the model’s predictions. It reveals that:
- True Positive (TP): 82 instances were correctly predicted as Class 1.
- True Negative (TN): 70 instances were correctly predicted as Class 0.
- False Positive (FP): 23 instances were incorrectly predicted as Class 1.
- False Negative (FN): 21 instances were incorrectly predicted as Class 0.
- The confusion matrix provides a detailed breakdown of the model’s predictions. It reveals that:
- Classification Report:
- Precision: Precision measures the accuracy of positive predictions. For Class 0, precision is 0.77, and for Class 1, precision is 0.78.
- Recall: Recall (Sensitivity or True Positive Rate) assesses the model’s ability to identify all relevant instances. Class 0 has a recall of 0.75, and Class 1 has a recall of 0.80.
- F1-Score: The F1-score is the harmonic mean of precision and recall. Class 0 has an F1-score of 0.76, and Class 1 has an F1-score of 0.79.
- Support: Indicates the number of actual instances for each class.
- Overall Summary:
- The weighted average accuracy across both classes is 0.78.
- The macro average, providing equal weight to both classes, for precision, recall, and F1-score is approximately 0.77.
- The weighted average, considering class imbalance, for precision, recall, and F1-score is also 0.78.
Step 7: MLflow
MLflow is an open-source platform designed to manage the end-to-end machine learning lifecycle. It provides tools and components to help with various stages of the machine-learning process, including experimentation, reproducibility, and deployment. MLflow was developed by Databricks but is now an open-source project with a community-driven development model.
Logging Experiments
MLflow logging refers to the process of recording and tracking various aspects of machine learning experiments using MLflow, an open-source platform for managing the end-to-end machine learning lifecycle. Logging in MLflow involves recording information such as parameters, metrics, artifacts, and model details during the execution of machine learning code. This logged information is crucial for reproducibility, collaboration, and model management.
Here are the key components of MLflow logging:
- Parameters:
- MLflow allows you to log the parameters used in your machine learning experiments. Parameters are the configuration settings or hyperparameters that influence the behavior of your models.
- Example:
mlflow.log_param("model_type", "Logistic Regression")
- Metrics:
- Metrics represent quantitative measurements of the performance of your models. Common metrics include accuracy, precision, recall, F1 score, etc.
- Example:
mlflow.log_metric("accuracy", accuracy)
- Artifacts:
- Artifacts are the output files or results generated during an experiment. This can include model files, plots, images, and any other relevant outputs.
- Model Logging:
- MLflow provides functions to log and save machine learning models in a standardized format. This allows you to easily reproduce and deploy models later.
- Example:
log_model(log_reg_model, "log_reg_model")
- Start and End Runs:
- MLflow uses the concept of runs to represent individual executions of your machine learning code. You can start a run using
mlflow.start_run()and end it usingmlflow.end_run().
- MLflow uses the concept of runs to represent individual executions of your machine learning code. You can start a run using
The following Python code demonstrates the integration of logistic regression with MLflow logging.
############################################
# STEP 7: ML-FLOW
############################################
import mlflow
from mlflow.sklearn import log_model
# Set the MLflow experiment name
experiment_name = 'Logistic Regression Heart Disease Prediction'
mlflow.set_experiment(experiment_name)
with mlflow.start_run():
# Create a logistic regression model
log_reg_model = LogisticRegression()
# Train the model on the transformed training features
log_reg_model.fit(X_train_transformed, y_train)
# Make predictions on the original test features
X_test_transformed = model.transform(X_test)
y_pred = log_reg_model.predict(X_test_transformed)
# Evaluate the model performance
accuracy = accuracy_score(y_test, y_pred)
conf_matrix = confusion_matrix(y_test, y_pred)
classification_rep = classification_report(y_test, y_pred)
# Log parameters
mlflow.log_param("model_type", "Logistic Regression")
mlflow.log_param("features_selected", len(selected_features))
mlflow.log_param("accuracy", accuracy)
# Log metrics
mlflow.log_metric("accuracy", accuracy)
# Log the model as an artifact
log_model(log_reg_model, "log_reg_model")Step 8: K-Fold Cross Validation
K-fold cross-validation is a technique used in machine learning to assess the performance of a predictive model. The dataset is split into “K” equally sized folds, and the model is trained and evaluated “K” times. During each iteration, one of the folds is used as the test set, and the remaining (K-1) folds are used as the training set. This process is repeated K times, with a different fold designated as the test set in each iteration.
The Python code below uses scikit-learn’s cross_val_score to perform 5-fold cross-validation on a logistic regression model (log_reg_model_cv) trained with transformed features (X_train_transformed), calculating accuracy scores for each fold and displaying both the individual scores and the mean cross-validation accuracy.
############################################
# STEP 8: K-FOLD CROSS VALIDATION
############################################
from sklearn.model_selection import cross_val_score
# Define the logistic regression model for cross-validation
log_reg_model_cv = LogisticRegression()
# Perform K-fold cross-validation (e.g., 5 folds)
cross_val_scores = cross_val_score(log_reg_model_cv, X_train_transformed, y_train, cv=5, scoring='accuracy')
# Display cross-validation results
print(f"Cross-Validation Accuracy Scores: {cross_val_scores}")
print(f"Mean Cross-Validation Accuracy: {np.mean(cross_val_scores):.2f}")
----------------- OUTPUT -----------------------
Cross-Validation Accuracy Scores: [0.76433121 0.80128205 0.80769231 0.76282051 0.82692308]
Mean Cross-Validation Accuracy: 0.79
------------------------------------------------The output shows the accuracy scores obtained from 5-fold cross-validation for a logistic regression model. The individual accuracy scores for each fold are displayed as an array: [0.764, 0.801, 0.808, 0.763, 0.827], representing the accuracy of the model on each subset of the training data.
The mean cross-validation accuracy, calculated by averaging these individual scores, is presented as 0.79. This mean accuracy provides an overall assessment of the model’s performance across different training and testing subsets, serving as a more reliable indicator of its generalization capabilities compared to a single train-test split. In this context, an accuracy of 0.79 suggests that, on average, the model correctly predicted the class labels for approximately 79% of the instances during the cross-validation process.
Step 9: Pre-Deployment
In this phase, the model is tested before deployment. This testing phase is crucial for identifying and addressing potential issues, bugs, or performance bottlenecks that could impact the reliability and functionality of the model in a live production environment.
unittest is a testing framework in Python that is part of the Python Standard Library. It provides a set of conventions and methods for writing and running tests to verify the correctness of code. The unittest module, inspired by Java’s JUnit, allows developers to create and execute test cases, organize tests into test suites, and perform various types of assertions to check whether the expected behavior of the code under test matches the actual behavior.
In the Python code below we use the unittest function to check if the output of the model is either 0 (no heart disease) or 1 (heart disease).
############################################
# STEP 9: UNITTEST
############################################
import unittest
class TestLogisticRegression(unittest.TestCase):
def setUp(self):
# Set up any necessary variables or configurations for the tests
pass
def tearDown(self):
# Clean up after the tests
pass
def test_predictions_binary(self):
# Ensure that predictions are either 0 or 1
valid_predictions = all(prediction in [0, 1] for prediction in y_pred)
self.assertTrue(valid_predictions, "Predictions should be either 0 or 1")
# Create a test suite
test_suite = unittest.TestLoader().loadTestsFromTestCase(TestLogisticRegression)
# Run the test suite
unittest.TextTestRunner().run(test_suite)
----------------- OUTPUT -----------------------
Ran 1 test in 0.001s
OK
------------------------------------------------The output indicates that the unit test ran successfully, and your test case (TestLogisticRegression) passed without any errors or failures.
Step 10: Deployment
Docker is a platform that enables developers to package and distribute applications, along with their dependencies, in a consistent and reproducible manner. In the context of machine learning (ML) deployment, Docker provides a way to encapsulate an ML model, its dependencies, and the runtime environment into a container.
To package the model using Docker, you’ll need to follow these general steps:
- Step 1: Create a Dockerfile: This file contains instructions for building a Docker image. In your case, it would include the necessary dependencies and setup for running your model. Create a file named
Dockerfile(no file extension) in the same directory as your Python script. Add the following content:
# Use an official Python runtime as a parent image
FROM python:3.8-slim
# Set the working directory to /app
WORKDIR /app
# Copy the current directory contents into the container at /app
COPY . /app
# Install any needed packages specified in requirements.txt
RUN pip install --no-cache-dir -r requirements.txt
# Make port 80 available to the world outside this container
EXPOSE 80
# Define environment variable
ENV NAME World
# Run app.py when the container launches
CMD ["python", "your_script_name.py"]- Step 2: Create a requirements.txt file: If you have external dependencies (e.g., specific versions of libraries), list them in a
requirements.txtfile. Create a file namedrequirements.txtand list the required packages:
pandas
matplotlib
numpy
seaborn
scikit-learn
mlflow- Step 3: Build the Docker image: Open a terminal, navigate to the directory containing your Dockerfile, and run the following command to build the Docker image:
docker build -t your_image_name .- Step 4: Run the Docker container: After the image is built, you can run a container based on that image. This command maps port 4000 on your host machine to port 80 in the Docker container. Adjust the ports as needed.
docker run -p 4000:80 your_image_nameThe model should now be packaged and running in a Docker container. This is a basic example, and depending on the actual requirements, we might need to customize the Dockerfile or take additional steps for a more complex setup.
Conclusion
In conclusion, this blog post walked through various crucial steps in the machine learning lifecycle, using a heart disease prediction task as an example. Here’s a summary of the key points covered:
- Step 1: Load & Explore Dataset:
- Import necessary libraries and load the heart disease dataset.
- Display basic information about the dataset.
- Visualize key aspects using Seaborn.
- Step 2: Exploratory Data Analysis (EDA):
- Understand the dataset’s main characteristics, patterns, and trends.
- Visualize distributions and correlations in the data.
- Step 3: Data Preparation:
- Clean and organize the data.
- Handle missing values and perform necessary transformations.
- Step 4: Feature Engineering:
- Normalize and standardize numerical features.
- Enhance the performance and interpretability of machine learning models.
- Step 5: Feature Selection:
- Use RandomForestClassifier for feature selection.
- Identify and focus on the most important features.
- Step 6: Model Training & Prediction:
- Train a logistic regression model.
- Evaluate model performance using accuracy, confusion matrix, and classification report.
- Step 7: MLflow Logging:
- Log experiments, parameters, metrics, and artifacts using MLflow.
- Facilitate reproducibility, collaboration, and model management.
- Step 8: K-Fold Cross Validation:
- Assess model performance using K-fold cross-validation.
- Obtain accuracy scores for different training and testing subsets.
- Step 9: Pre-Deployment Testing:
- Use unittest to perform unit testing.
- Check the validity of model predictions.
- Step 10: Deployment with Docker:
- Package the model, dependencies, and runtime environment into a Docker container.
- Create a Dockerfile and build the Docker image.
- Run the Docker container for deployment.
By following these steps, from data exploration to deployment, we have covered essential aspects of an end-to-end machine learning project.